Ever wanted to use your spare time in EVE online tackling some real-world scientific problems? Maybe earn ISK or some other reward for your effort? Look no further; Project Discovery is coming soon.
Project Discovery Brings Citizen Science to New Eden
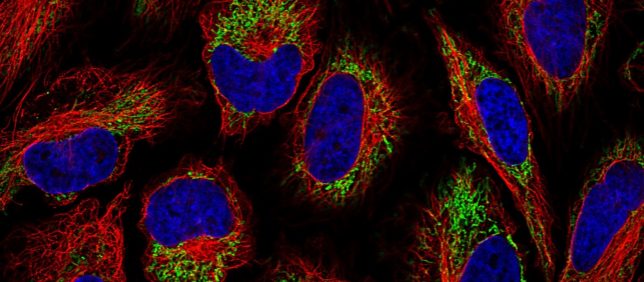
Like other online Citizen Science projects such as folding@home, Project Discovery will put real world biomedical data into the hands of EVE players for individual analysis. After some light training by analyzing data that has already been validated to some extent, players will be tasked data within EVE that has not be entirely explained. Together, players will reach a statistically accurate consensus, and the downstream data can be tackled by more in-depth experimentation by researchers worldwide. Besides, who doesn’t like looking at beautiful fluorescent confocal images of cells?
As announced by CCP Scarpia:
“Together with Swiss startup company Massively Multiplayer Online Science and Reykjavík University, CCP has been working on bringing real world scientific research into the EVE game experience. For the launch of Project Discovery we are collaborating with The Human Protein Atlas (HPA), a Swedish research group that builds and provides a public database of human proteins that gets used by thousands of medical research groups. By participating in Project Discovery you will be helping HPA to analyze new protein images to expand their database and in turn contributing to the wellbeing of future patients being treated around the world.”
A starting point at launch
The emphasis of Project Discovery at launch will be based around the sub-cellular spatial localization of proteins in human cell lines to cellular components.
Human cell lines are cells that were isolated from a specific patient tissue by biopsy or dissection that are then ‘transformed’ by various specific factors, or spontaneously, to divide indefinitely similar to the behavior of cancer cells, all the while cultured in a dish. Obviously these cells do not mimic the in vivo (or inside a human) condition, as cells in the human body besides cancer cells, and possibly niche stem cell populations, are not known to divide indefinitely.
Cell lines can be derived from different tissues to bring about different varieties of lines. Cell lines in a sense act as an excellent starting point for further analysis being a functional biological bag containing organelles and many other biological machinery. A big part of the first incarnation of project discovery will be in determining what cellular machinery specific proteins localize to.
The Human Protein Atlas also contains data generated from actual human tissue (and therefore “real” in vivo cells), although it is not clear at this time what levels of this atlas will be used in Project Discovery.
What EVE Players Should Expect
As most eloquently stated by CCP Scarpia,
“Project Discovery is a classified research program founded and run by the Sisters of EVE. Their project lead, Professor Lundberg, will recruit you and provide you with a basic tutorial to be able to recognize specific patterns in biological samples already classified by experts. As soon as you graduate from the training program, you and other graduated contributors will be tasked with analyzing unique images fresh from the lab.
To start you off you are tasked with increasingly difficult tasks which already have a solution, when you have finished with those you will get tasks that don‘t have a solution. Once enough contributors have reached a statistically accurate consensus on a task which has no solution, you will receive a boost to your Project Discovery accuracy rating. For every task you solve, the SoE will reward you with ISK and LP, as well as Analysis Points which contribute to your Project Discovery rank.”
How to Get In
Players can access Project Discovery on the Singularity test server via the neocom (under ‘Business’) as seen here. Photo credit goes to /u/thesared, who posted the images to the EVE subreddit.
Another user, Erutor, posted this amusing hypothetical scenario.
“What are you doing on your computer, Timmy? It is 3AM.”
“I’m working with the Human Protein Atlas research project in Sweden to identify all proteins our genes are coding for, and their spatial pattern of expression, to ultimately understand their function and connection to disease, Mom.”
“Oh, ok, I thought you were playing a game. Don’t study too late.”
“Yes, ma’am.”
Project Discovery is currently in initial testing phases on the EVE test server Singularity. A mass test is scheduled for Jan 28. The original devblog post can be found here. Background material on proteins and the science behind Project Discovery can be found on the next page. Readers interested in science and biology are encouraged to take a look!
Are you looking forward to the addition of this feature to EVE online? Let us know your perspective in the comments below.

What are proteins, anyways?
Proteins can be thought of as the primary workers and scaffolding of the cellular environment, providing critical structure and function to cells. Proteins are synthesized and may remain within the cellular environment for their entire life (of the protein) being embedded within cellular membranes, translocated to specific organelles, or existing only in the cytosol, the part of the cytoplasm that is not held by any of the organelles in the cell. Many others proteins are actually secreted outside the cell after synthesis and may act on the extracellular environment. Indeed a specific type of protein could possibly be found within the cell and outside the cell at the same time. Many proteins are multifunctional and act in a context specific manner or require some co-requisite. Therefore, the localization data presented is only a snapshot, and any further conclusions must be made by downstream experimentation.
In the human genome, there are over 20,000 protein coding genes that can actually be rearranged to form many other products. DNA found in the nucleus or the mitochondria is transcribed into RNA, which is then processed into messenger RNA, that is in turn translated into proteins by ribosomes. Think of the DNA as an original blueprint that no one touches, RNA as the blueprint copies, and ribosomes as the production lines.
Proteins are then ‘folded’ into some specific form by local thermodynamics, other proteins, bonding events, and other factors. Proteins can further link-up into multi-protein scaffolds that are functionally unique, containing multiple different proteins, or may form aggregate clusters containing various cellular components. There could be tens of thousands or more unique proteins expressed by the various cells of the body. Damaged or unnecessary proteins are targeted for recycling to various cellular systems including the proteasomal system (similar to a protein blender) or by mechanisms such as autophagy.
A large proportion of the human genome actually does not even encode proteins. This DNA is regulatory, altering the ability of specific sequences to be expressed, and in some cases being transcribed into regulatory RNAs that do not directly translate into proteins, but may alter the expression of proteins. Some regulatory RNAs can even shutdown an entire chromosome, as is the case for the long non-coding RNA Xist that turns off one X chromosome in female mammals, most vividly seen in calico cats.
How can proteins be tracked?
The two biggest methods that are being employed by The Human Protein Atlas for protein localization and detection are by gene-fusion and immunostaining.
By gene-fusion, a fluorescent protein coding DNA sequence isolated from coral or jellyfish, such as green fluorescent protein for instance, is placed at one end of a protein coding sequence. Once expressed and translated, the protein will contain a fluorescent ‘tag’ that can be seen through microscopes. This has consequences, as the tags can cause the proteins to exhibit altered patterns and states.
By immunostaining, antibodies are generated in mice, rabbits, donkeys or chickens after injection with specific proteins, usually produced in e. coli or some vertebrate cell lines, and then further isolated and purified. These antibodies are then added to fixed (no longer alive) and permeabilized cells that will then bind their specific protein target. A secondary antibody, which targets species specific antibodies, is then added, containing a fluorescent tag. A major limitation of antibodies is non-specific binding and usually antibodies are validated, although not always, by some other method of experimentation.
By reducing the amount of protein being made by the cell, inferences can be made into the specificity of any one protein labeling technique. One method is by siRNA-mediated gene silencing. This involves synthetic RNA being added to cells to reduce the levels of messenger RNA made, and therefore the amount of protein produced by the cell.
Once it is known where a protein localized, researchers can mutate the coding sequence to try and find the exact amino acid requirements for this localization phenomena. Once those parameters are found, other factors such as effector proteins or cellular states contributing to the researched protein can be further explored.
What the analysis leaves out
In different cells of the body, sometimes proteins are expressed either at different levels, being hard to detect from one cell type to the next. In addition, many proteins undergo modifications after synthesis that alter their specific functions and localization in the context of any one specific biological phenomena. This sort of information will not be immediately identified by just microscopic localization data alone.
Proteins aren’t the only important part of a cell. Carbohydrates and lipids that are produced as a consequence of enzymatic processes or via the recycling of cellular components (either from food or self) are equally important, although very elusive in direct study. This isn’t necessarily a limitation of Project Discovery, but a major void in the current era of biomedical research.
Why Project Discovery is so Important
Biomedical experiments that generate microscopic images are difficult to analyze in bulk by computational means (although not impossible) and must be analyzed by people in real-time that can bring about its own biases. These experiments also generate an enormous amount of data, like many other biomedical techniques, that further encumbers the process.
However, the analysis of these images is really not too difficult, and getting a large body of people to assist in this process is really helpful for generating non-biased hypotheses. More people providing interpretation of imaging data provides less bias to any one rationalization, providing a greater reliability in the conclusions made.
It is entirely possible that, through the combined efforts of capsuleers, more data can be interpreted in an unbiased way. Mankind’s body of biomedical knowledge will grow, along with our ISK wallets.